mpoly is the most basic function used to create objects of class mpoly.
as.mpoly(x, ...)
Arguments
| x | an object |
|---|---|
| ... | additional arguments to pass to methods |
Value
the object formated as a mpoly object.
See also
Examples
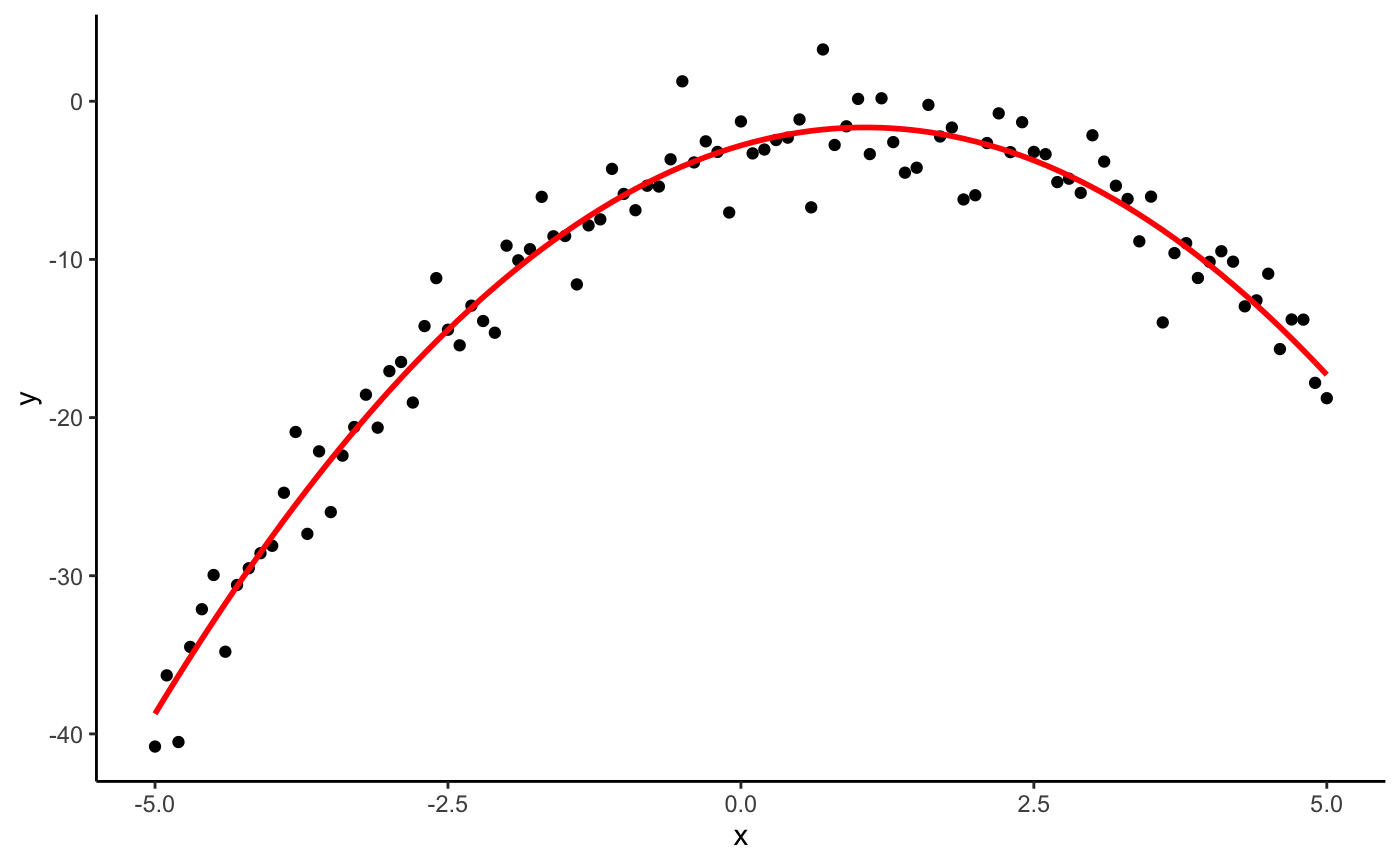
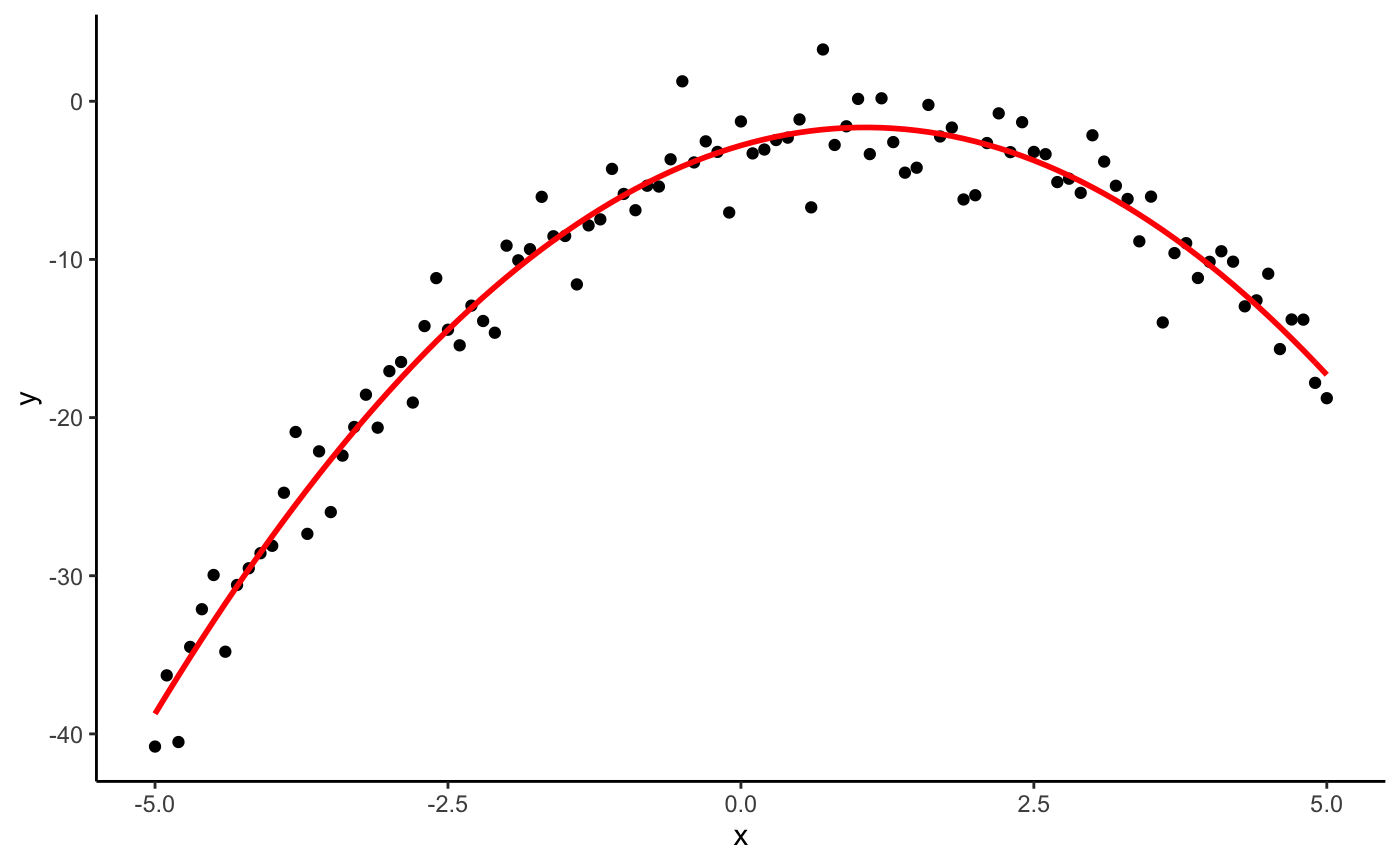
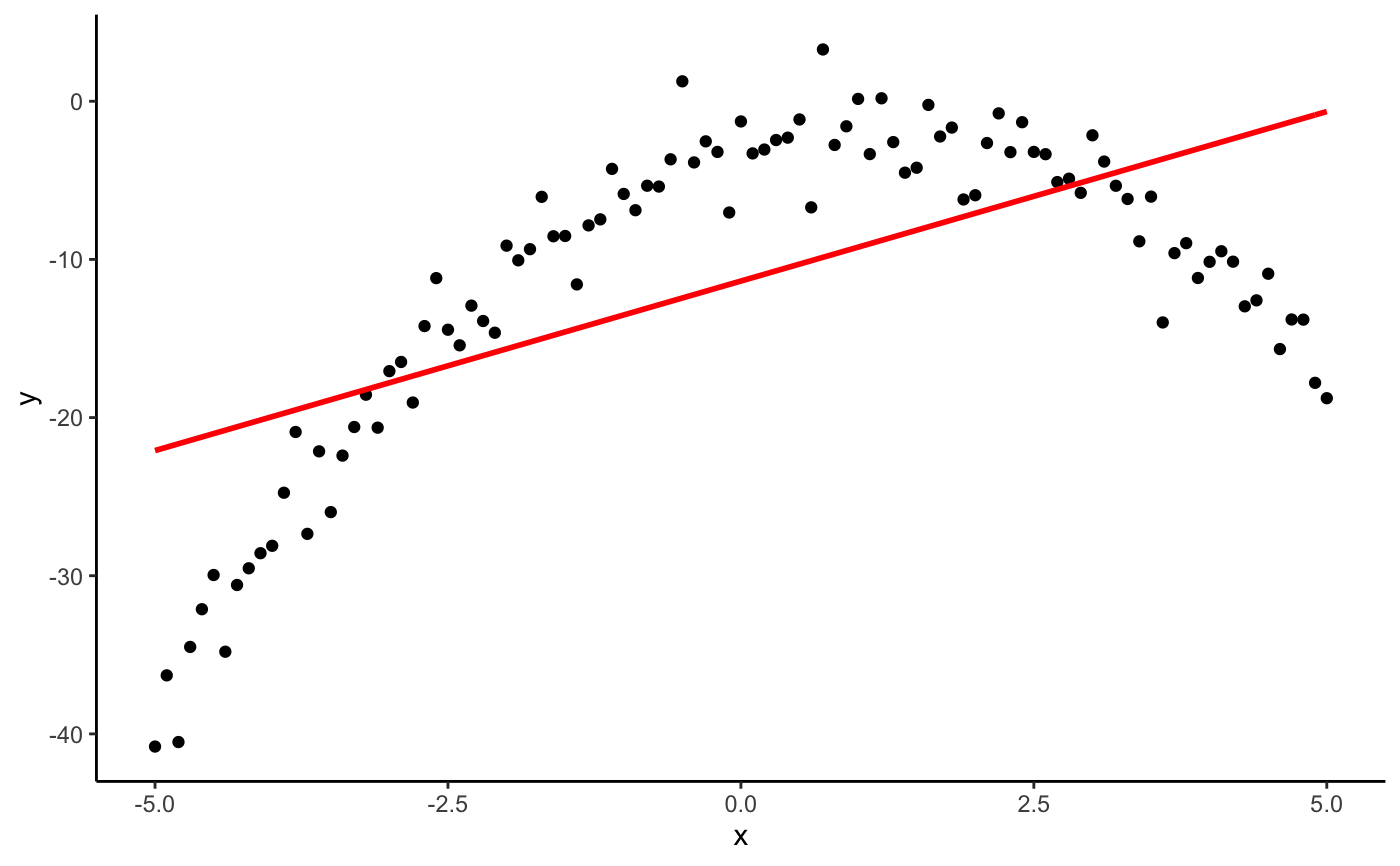
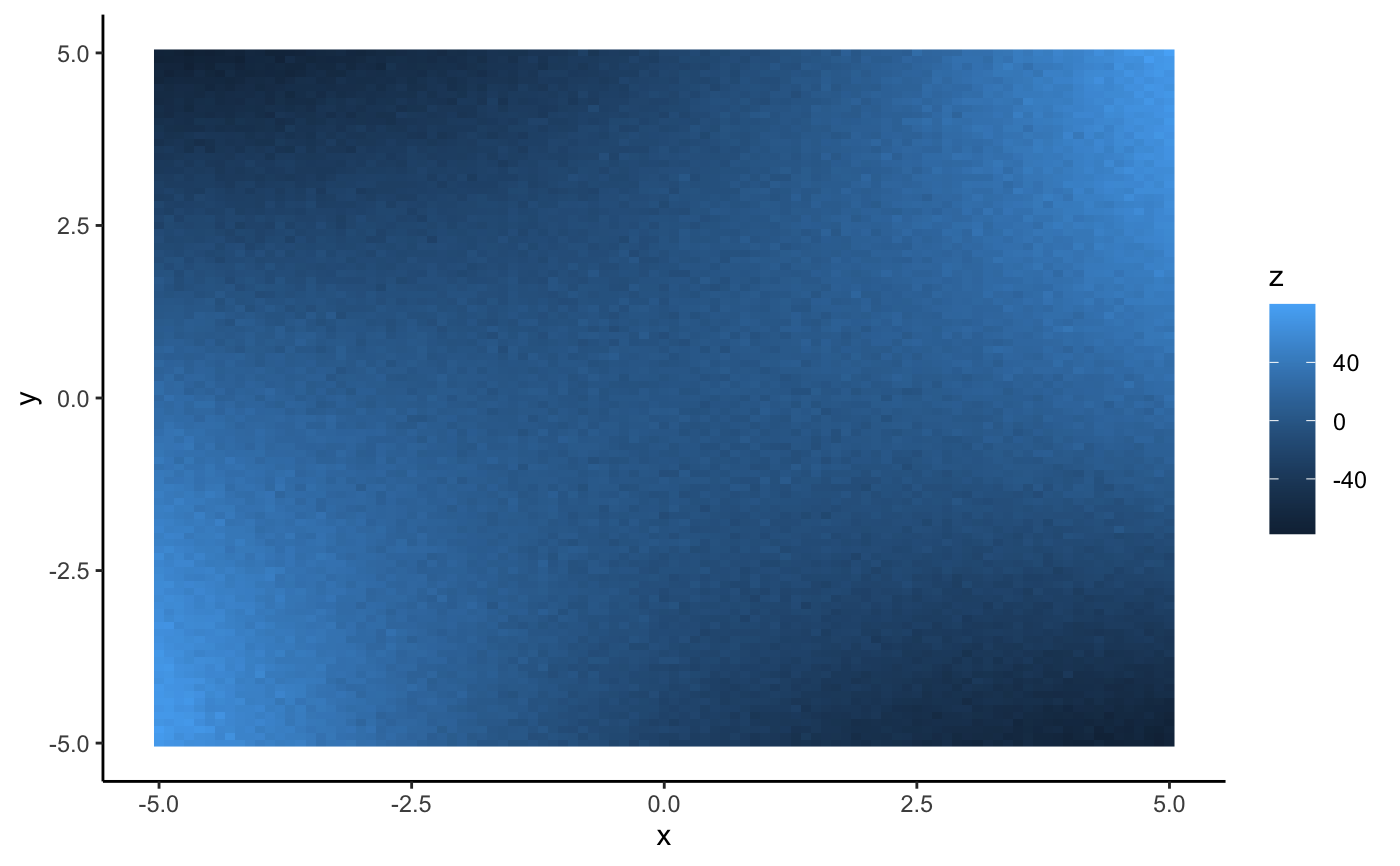
#> #>#> #> #>library(dplyr)#> #>#> #> #>#> #> #>#> #> #>n <- 101 s <- seq(-5, 5, length.out = n) # one dimensional case df <- data.frame(x = seq(-5, 5, length.out = n)) %>% mutate(y = -x^2 + 2*x - 3 + rnorm(n, 0, 2)) (mod <- lm(y ~ x + I(x^2), data = df))#> #> Call: #> lm(formula = y ~ x + I(x^2), data = df) #> #> Coefficients: #> (Intercept) x I(x^2) #> -2.794 2.144 -1.009 #>(p <- as.mpoly(mod))#> 2.143584 x - 1.008662 x^2 - 2.793865#>#> #> Call: #> lm(formula = y ~ poly(x, 2, raw = TRUE), data = df) #> #> Coefficients: #> (Intercept) poly(x, 2, raw = TRUE)1 poly(x, 2, raw = TRUE)2 #> -2.794 2.144 -1.009 #>(p <- as.mpoly(mod))#> 2.143584 x - 1.008662 x^2 - 2.793865#>#> #> Call: #> lm(formula = y ~ poly(x, 1, raw = TRUE), data = df) #> #> Coefficients: #> (Intercept) poly(x, 1, raw = TRUE) #> -11.367 2.144 #>(p <- as.mpoly(mod))#> 2.143584 x - 11.36749#># two dimensional case with ggplot2 df <- expand.grid(x = s, y = s) %>% mutate(z = x^2 - y^2 + 3*x*y + rnorm(n^2, 0, 3)) qplot(x, y, data = df, geom = "raster", fill = z)#> #> Call: #> lm(formula = z ~ x + y + I(x^2) + I(y^2) + I(x * y), data = df) #> #> Coefficients: #> (Intercept) x y I(x^2) I(y^2) I(x * y) #> 0.062597 -0.002988 -0.007926 0.997782 -1.005816 2.995417 #>#> #> Call: #> lm(formula = z ~ poly(x, y, degree = 2, raw = TRUE), data = df) #> #> Coefficients: #> (Intercept) poly(x, y, degree = 2, raw = TRUE)1.0 #> 0.062597 -0.002988 #> poly(x, y, degree = 2, raw = TRUE)2.0 poly(x, y, degree = 2, raw = TRUE)0.1 #> 0.997782 -0.007926 #> poly(x, y, degree = 2, raw = TRUE)1.1 poly(x, y, degree = 2, raw = TRUE)0.2 #> 2.995417 -1.005816 #>(p <- as.mpoly(mod))#> -0.00298808 x + 0.997782 x^2 - 0.007925533 y + 2.995417 x y - 1.005816 y^2 + 0.06259731#>